Exploring crop health and its associations with fungal soil microbiome composition using machine learning applied to remote sensing data
Από RemoteSensing Wiki
| (3 ενδιάμεσες αναθεωρήσεις δεν εμφανίζονται.) | |||
| Γραμμή 4: | Γραμμή 4: | ||
'''Source:''' https://doi.org/10.1038/s43247-025-02330-0 <br/> | '''Source:''' https://doi.org/10.1038/s43247-025-02330-0 <br/> | ||
'''Date: 7 May 2025''' <br/> | '''Date: 7 May 2025''' <br/> | ||
| + | |||
| + | [[Αρχείο: MODEL.png | thumb | '''Fig 1.''' Model comparisons for the linear regression model (Linear) and random forest model (RF) using the root mean squared error (RMSE) for the test set (20%) and the full dataset (including the test set). Note that the R2 indicates the explained variance for the full dataset including the test set.]] | ||
| + | |||
| + | [[Αρχείο: BOXPLOTS.png | thumb | '''Fig 2.'''Boxplots of the residual NDVI values from each RF model of Table 2 within clusters 1 and 2 originating from the unsupervised clustering of the biodiversity samples.]] | ||
| + | |||
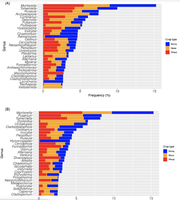
| + | [[Αρχείο: GENERA.png | thumb | '''Fig 3.''' A Visualization of the 30 most abundant taxonomies per crop for the first cluster. B Visualization of the 30 most abundant taxonomies per crop for the second cluster.]] | ||
<u> '''Summary and Introduction''' </u> <br/> | <u> '''Summary and Introduction''' </u> <br/> | ||
| Γραμμή 37: | Γραμμή 43: | ||
For the discussion section, the researchers compared their findings to the findings of past literature and explained any possible limitations of their models. Some areas of the results that agreed with the literature are the following: NDVI is affected by the seasons, observing a maximum NDVI around harvest months. Additionally, soil carbon content has a non-linear correlation with NDVI. Furthermore, meteorological variables such as temperatures and soil moisture have the highest importance with NDVI. Lastly, the researchers acknowledge that their results confirm past results regarding their abiotic model to remove non-living influence from the NDVI response. | For the discussion section, the researchers compared their findings to the findings of past literature and explained any possible limitations of their models. Some areas of the results that agreed with the literature are the following: NDVI is affected by the seasons, observing a maximum NDVI around harvest months. Additionally, soil carbon content has a non-linear correlation with NDVI. Furthermore, meteorological variables such as temperatures and soil moisture have the highest importance with NDVI. Lastly, the researchers acknowledge that their results confirm past results regarding their abiotic model to remove non-living influence from the NDVI response. | ||
There were several limitations to the study that were addressed by the researchers. First, their meteorological data is confined to certain ranges, such as temperature ranges between 280K and 290K. This means that their abiotic model cannot handle extreme weather well. Additionally, biological data were only obtained from 2018, so any changes associated with time were not properly observed. For future studies, using larger geospatial data and larger time periods may benefit research. Also, using satellites with high resolution may provide noteworthy results since they may be able to map biotic, living effects better. Researchers also address that testing fungal interactions both in vivo and in vitro may be useful. The example they give is with the Mortierella, and how there have been no known field experiments on how different levels of the fungus can affect the NDVI. They then give an idea of how such experiments could be carried out. Another potential candidate for future research in the field is the use of species-level taxonomy for NDVI data, since this particular paper only used genus-level taxonomy, which is broader. | There were several limitations to the study that were addressed by the researchers. First, their meteorological data is confined to certain ranges, such as temperature ranges between 280K and 290K. This means that their abiotic model cannot handle extreme weather well. Additionally, biological data were only obtained from 2018, so any changes associated with time were not properly observed. For future studies, using larger geospatial data and larger time periods may benefit research. Also, using satellites with high resolution may provide noteworthy results since they may be able to map biotic, living effects better. Researchers also address that testing fungal interactions both in vivo and in vitro may be useful. The example they give is with the Mortierella, and how there have been no known field experiments on how different levels of the fungus can affect the NDVI. They then give an idea of how such experiments could be carried out. Another potential candidate for future research in the field is the use of species-level taxonomy for NDVI data, since this particular paper only used genus-level taxonomy, which is broader. | ||
| + | |||
| + | [[category:Οικολογία]] | ||
Παρούσα αναθεώρηση της 12:18, 13 Ιανουαρίου 2026
Article title: "Exploring crop health and its associations with fungal soil microbiome composition using machine learning applied to remote sensing data"
Authors: Mathies Brinks Sørensen, David Faurdal, Giovanni Schiesaro, Emil Damgaard Jensen, Michael Krogh Jensen, Line Katrine Harder Clemmensen
Source: https://doi.org/10.1038/s43247-025-02330-0
Date: 7 May 2025

Summary and Introduction
The goal of this study was to examine how remote sensing combined with machine learning could assist in understanding crop health. One of the largest growing concerns around the globe has been an increase in food insecurity as populations continue to rise. For this reason, it is important to apply sustainable agricultural practices in order to increase crop yields and productivity.
Smart farming methods integrate remote sensing technologies such as satellite imagery and/or drones with data analytics tools to monitor fields. The common satellites in such applications are Sentinel-2 and Landsat 8, which have medium resolution but only sample every several days. The MODIS satellite is another option since it provides daily samples; however, it is low resolution and provides less accurate analyses. Multispectral images from these satellites are used to calculate vegetation indices that are used to visualize vegetation characteristics. The most common vegetation indices are the normalized difference vegetation index (NDVI), the enhanced vegetation index (EVI), and the leaf area index (LAI).
Of all the vegetation indices, NDVI is the most popular because of the large number of factors that can affect it. Some of these include soil moisture, climate, nutrients, crop type, and temperatures. This paper conducted a literature review on how different microbial species may affect the NDVI. Prior research has proven a link between certain bacteria and NDVI values. As of now, there has been limited research linking the existence of certain fungi and NDVI values. Though the research that does exist has shown that soil decomposers play a role in higher NDVI values and that fungal richness is connected to environmental factors such as pH and landscape types. The lack of research on fungal biomes motivated this study to better examine the connection between fungal soil composition and NDVI values.
Methods and Data
The first step of the methodology was to download the satellite images and apply the NDVI. Low NDVI depicts bad crop health, while high NDVI depicts good crop health. Then, the NDVI values were adjusted for abiotic (non-living) factors by removing their influence through a random forest model. Next, the NDVI values were analyzed based on different fungal soil microbiomes.
The abiotic data, which were used to adjust the NDVI, came from various sources. Topsoil composition data were obtained from the LUCAS 2018 topsoil dataset, but only the three most prevalent crop types were used: wheat, barley, and maize. The data was further filtered by removing instances during winter months since most of the images were covered in snow. Climate data came from the ERA5 Copernicus dataset, and information related to soil properties was obtained, such as soil temperature, soil moisture, soil type, and air temperature. These variables were then linked to each NDVI observation. This step was vital to make sure that any recorded changes in the NDVI values resulted exclusively from fungi and not any other non-living factors. If too many parameters were changing simultaneously without adjustments, no reasonable correlation could be examined. The last dataset used was the LUCAS biodiversity dataset. This dataset contains a total of 885 biosamples, of which 115 were for wheat, barley, and maize crops, during the specific time period.
Results
The first results of this paper demonstrated which model, linear regression or random forest (RF), predicted the NDVI values better. As shown in Table 1, the RF model outperformed the linear regression model since it had higher R^2 values and lower RMSE values. The higher R^2 values indicate a greater explained variance. These models used 16 parameters to do the calculations, obtained from the data sets described in the methods section.
Since the RF models outperformed the linear regression model, they were used to carry out the rest of the fungal soil modeling. Two major clusters of fungal genera were identified through unsupervised clustering based on statistical groupings of their properties. Based on the boxplots from Figure 2, it is clear that the residual NDVI values were higher for cluster 1 than cluster 2, indicating better plant health.
To gain a better understanding of which fungi may be contributing to the NDVI values, the frequency of the fungal genera was visualized in Fig. 3. The researchers explain that fungi such as Tomentella and Mortierella, which typically are associated with plant health, are more abundant in (wheat) cluster 1. ** Based on my own observation, however, it seems that both clusters have very similar frequency percentages of Mortierella so I fail to see how it is an indication of plant health. Fusarium, on the other hand, was very prevalent in cluster 2, while not appearing at all in the top 30 of cluster 2 genera. This may indicate that Fusarium is a pathogenic genus, contributing to poorer plant health and, consequently, lower NDVI values. A bootstrap resampling approach helped identify 99 and 114 genera in clusters 1 and 2, respectively, as non-outlier data.
Discussion
For the discussion section, the researchers compared their findings to the findings of past literature and explained any possible limitations of their models. Some areas of the results that agreed with the literature are the following: NDVI is affected by the seasons, observing a maximum NDVI around harvest months. Additionally, soil carbon content has a non-linear correlation with NDVI. Furthermore, meteorological variables such as temperatures and soil moisture have the highest importance with NDVI. Lastly, the researchers acknowledge that their results confirm past results regarding their abiotic model to remove non-living influence from the NDVI response.
There were several limitations to the study that were addressed by the researchers. First, their meteorological data is confined to certain ranges, such as temperature ranges between 280K and 290K. This means that their abiotic model cannot handle extreme weather well. Additionally, biological data were only obtained from 2018, so any changes associated with time were not properly observed. For future studies, using larger geospatial data and larger time periods may benefit research. Also, using satellites with high resolution may provide noteworthy results since they may be able to map biotic, living effects better. Researchers also address that testing fungal interactions both in vivo and in vitro may be useful. The example they give is with the Mortierella, and how there have been no known field experiments on how different levels of the fungus can affect the NDVI. They then give an idea of how such experiments could be carried out. Another potential candidate for future research in the field is the use of species-level taxonomy for NDVI data, since this particular paper only used genus-level taxonomy, which is broader.